Cancer Biology & Genetics Program
The Christine Mayr Lab
Research

Professor
My laboratory studies the regulatory and structural roles of mRNAs in the cytoplasm. Specifically, we investigate how 3′UTRs regulate protein functions in a manner that is independent of protein abundance.
We found that inclusion or exclusion of the 3′UTR of an mRNA that encodes a transcription factor did not change how much of the transcription factor protein was expressed but determined the location of protein synthesis of the transcription factor as well as its induced gene expression program. Moreover, we observed that expression of a chromatin regulator from an mRNA with or without its 3′UTR resulted in similar expression of the protein in the nucleus but changed the enzymatic activity of the chromatin regulator in the nucleus.
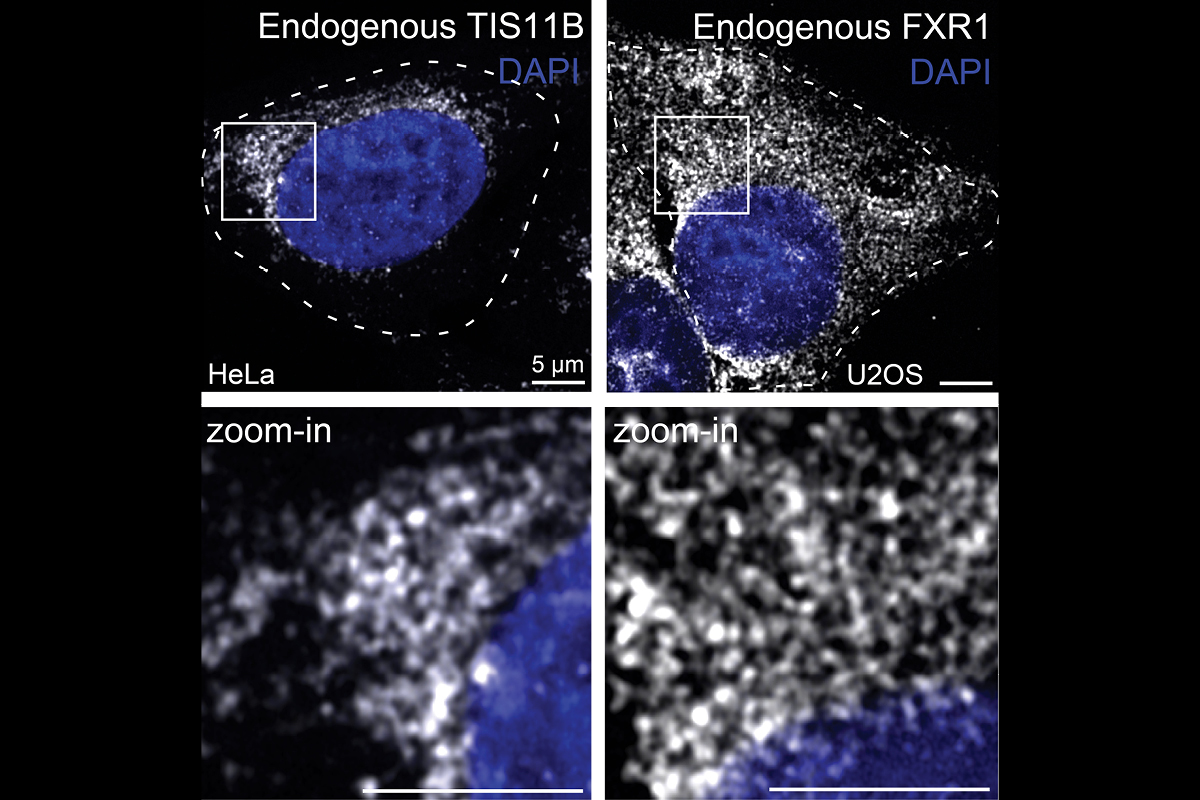
These examples illustrate that in addition to the coding sequence (which is translated into protein), other parts of mRNAs, specifically the 3′ untranslated region (3′UTR), can control protein activity and protein function in a manner that is independent of protein abundance. We are currently studying the mechanism by which 3′UTRs control protein activity. In the course of these studies, we found that 3′UTRs determine where in the cytoplasm an mRNA transcript is translated. Moreover, we found that the cytoplasm is highly compartmentalized into different translation environments. We identified several mRNA-based translation environments and have evidence that within these environments, mRNAs play major regulatory roles. We currently study the three mRNA-based translation environments that were discovered by us. They include TIS granules, the FXR1 network, and a poorly characterized environment generated by TIAL1 on the endoplasmic reticulum.
We are interested in the following questions:
- What is the mechanism by which translation in different mRNA-based environments controls protein activity in the nucleus?
- What is the contribution of 3′UTRs to cellular phenotypes, with a focus on transcription factor and chromatin regulator functions during iPS cell differentiation?
- What is the relationship between mRNAs and intrinsically disordered regions (IDRs)? This is based on our findings that mRNAs can act as chaperones for IDRs and can change their conformational states.
- What is the molecular code by which different translation environments control transcription factor and chromatin regulator functions?
- Can we develop novel RNA therapeutics to activate or deactivate transcription factors or even target the separate functions of multifunctional transcriptional regulators?
Research Projects
Featured News



Publications Highlights
Chen, X., Fansler, M.M., Janjoš, U., Ule, J., and Mayr, C. (2024). The FXR1 network acts as signaling scaffold for actomyosin remodeling. Cell, in press. Also on bioRxiv, 2023.2011.2005.565677.
Horste, E.L., Fansler, M.M., Cai, T., Chen, X., Mitschka, S., Zhen, G., Lee, F.C.Y., Ule, J., and Mayr, C. (2023). Subcytoplasmic location of translation controls protein output. Mol Cell 83, 4509-4523.e4511.
Ma W, Zhen G, Xie W, Mayr C. In vivo reconstitution finds multivalent RNA-RNA interactions as drivers of mesh-like condensates. eLife, 10:e64252 (2021). PMID: 33650968
Ma W, Mayr C. A membraneless organelle associated with the endoplasmic reticulum enables 3′UTR-mediated protein-protein interactions. Cell 175, 1492-1506 (2018). PMID: 30449617
Berkovits BD, Mayr C. Alternative 3’UTRs act as scaffolds to regulate membrane protein localization. Nature 522, 363-367 (2015). Epub 2015 Apr 20. PMID:25896326
People

Christine Mayr, MD, PhD
Professor
- Molecular and cell biologist Christine Mayr studies how 3′UTRs regulate protein functions and how mRNAs contribute to cytoplasmic organization.
- MD, Free University Berlin
- PhD, Humboldt University Berlin
- mayrc@mskcc.org
- Email Address
- Nadine Hernández
- Office Phone
- Download CV
- PDF File
Members








Research Associate

Research Scholar

Research Scholar

Research Technician

Senior Administrative Assistant

Postdoctoral Fellow

Graduate Student

Graduate Student


- BS, University of Michigan Ann Arbor








Lab Alumni

Postdoctoral Fellow

Graduate Student

Graduate Student, Tri-I CBM Program

Postdoctoral Fellow

Postdoctoral Fellow

Scientist/Payload Engineer, Apertura Gene Therapy, New York, NY

Master Student

Postdoctoral Fellow

Research Technician

Postdoctoral Fellow

Research Technician

Postdoctoral Fellow

Research Technician

Senior Research Scientist

Research technician

Postdoctoral Fellow

Research Technician

Postdoctoral Fellow

Administrative Assistant

Administrative Assistant

Postdoctoral Fellow
Lab Affiliations
Achievements
- Luise and Allston Boyer Young Investigator Award for Basic Research (2019)
- NIH Director’s Pioneer Award (2016)
- Pershing Square Sohn Prize for Young Investigators in Cancer Research (2015)
- Damon Runyon-Rachleff Innovation Award, Damon Runyon Cancer Research Foundation (2012)
- Selected as ‘Cell Scientist to watch’ by Journal of Cell Science (2015)
Read more
- Science Signaling Breakthrough of the year (2013)
Open Positions
To learn more about available postdoctoral opportunities, please visit our Career Center
To learn more about compensation and benefits for postdoctoral researchers at MSK, please visit Resources for Postdocs
Career Opportunities
Get in Touch
-
Lab Head Email
-
Office Phone
Disclosures
Doctors and faculty members often work with pharmaceutical, device, biotechnology, and life sciences companies, and other organizations outside of MSK, to find safe and effective cancer treatments, to improve patient care, and to educate the health care community.
MSK requires doctors and faculty members to report (“disclose”) the relationships and financial interests they have with external entities. As a commitment to transparency with our community, we make that information available to the public.
Christine Mayr discloses the following relationships and financial interests:
No disclosures meeting criteria for time period
The information published here is a complement to other publicly reported data and is for a specific annual disclosure period. There may be differences between information on this and other public sites as a result of different reporting periods and/or the various ways relationships and financial interests are categorized by organizations that publish such data.
This page and data include information for a specific MSK annual disclosure period (January 1, 2023 through disclosure submission in spring 2024). This data reflects interests that may or may not still exist. This data is updated annually.
Learn more about MSK’s COI policies here. For questions regarding MSK’s COI-related policies and procedures, email MSK’s Compliance Office at ecoi@mskcc.org.
