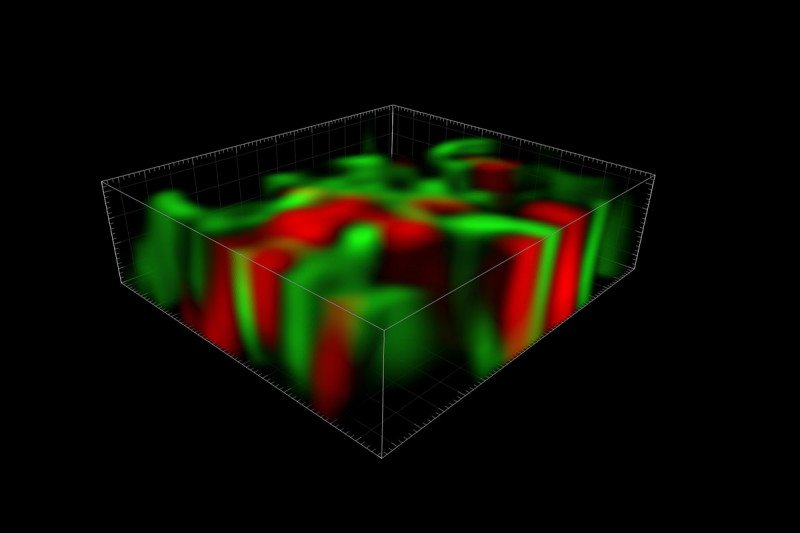
A three-dimensional reconstruction of the TIGER domain, a newly discovered organelle involved in protein synthesis.
Many people probably remember cell organelles from high school biology. Mitochondria are a cell’s “energy factories.” The nucleus stores genetic information. The Golgi apparatus packages proteins for distribution.
There haven’t been many changes to this roster of cell parts since it was assembled more than a century ago. But scientists at the Sloan Kettering Institute announced today that they have found a new organelle to add to the list.
They call it the TIGER domain.
This fearsome-sounding space occupies a large portion of the cell and is closely allied with the site where proteins are made. According to Christine Mayr, one of two cell biologists who discovered it, the TIGER domain is where messenger RNAs (mRNAs) encoding certain kinds of proteins find the appropriate environment — lair? — in which to grow up.
“It’s really a sorting mechanism,” Dr. Mayr says. “The organelle admits specific messenger RNAs according to certain rules and excludes others. Then it shapes how the proteins made from those messenger RNAs will function.”
In a new paper published today in Cell, she and Weirui Ma, a postdoctoral fellow in her lab, discuss what they have learned so far about the newly discovered organelle and how they found it. Like other tigers, it was hiding in plain sight.
A Place for Protein Interactions
The first three letters of TIGER stand for TIS granules; ER is for endoplasmic reticulum. TIS granules are a network of interconnected proteins that bind RNA. ER is where protein synthesis occurs. The combination of TIS granules plus ER creates a distinct space where mRNA and proteins can collect and interact.
Dr. Ma named this space TIGER not only because the acronym fits but also because the striped pattern of the TIS granules interweaving with the ER resembles the orange-and-black coloring of a tiger. The scientists have found the TIGER domain in every cell type they’ve looked at so far.
You might say the domain provides a kind of secret sauce for protein interactions.
“Most people who study this topic think that if two proteins are present in the same area of a cell and bump into each other, that’s enough for them to interact,” Dr. Mayr says. “We’ve found that’s not the case. Some protein interactions can take place in the TIGER domain and nowhere else.”
Take the example of a newly synthesized membrane protein destined for the surface of a cell. It needs to interact with several other proteins to complete its journey from the ER to the cell surface. Even though these particular proteins are distributed throughout the cell’s watery cytoplasm, they can only mix successfully when they are inside the TIGER domain.
Remove the TIGER domain from the cell and the membrane protein never becomes equipped to reach the cell surface.
Deciphering the TIGER Postal Code
Drs. Mayr and Ma didn’t set out to discover a new organelle. They stumbled upon it in the course of researching another topic: 3’UTRs (3 prime untranslated regions). These are parts of an mRNA molecule that do not contain protein-coding information and are not translated into a protein. They sometimes have strings of nucleotide letters called AU-rich elements (AREs). RNA-binding proteins attach to these AREs and influence what happens to the mRNAs.

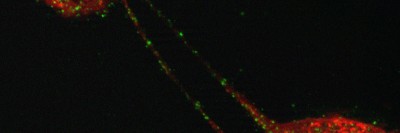
A fluorescent microscopy image of the TIGER domain, with TIS granules shown in red and the ER shown in green. The central black hole is the nucleus.
The investigators were interested in an RNA-binding protein called TIS11B. They gave it a fluorescent tag and then looked through a microscope at its distribution. A large region of the cell next to and interwoven with the ER lit up. This specific location suggested to them that TIS11B was somehow involved in protein synthesis.
Drs. Mayr and Ma then noticed something else: The number of AREs in an mRNA correlated with whether it was located inside or outside of this region. MRNAs with many AREs congregated in this region, while mRNAs with few or no AREs did not.
This led them to hypothesize that perhaps the number of AREs provides a kind of postal code for mRNAs. To test this idea, they performed an experiment. They systematically altered the number of AREs in several mRNAs and watched where those mRNAs went. Sure enough, the presence of a large number of AREs determined whether an mRNA found its way to the area that would come to be called TIGER.
Organelle Without Borders, or How TIGER Earned Its Stripes
Unlike other well-known organelles — say the nucleus or mitochondria — the TIGER domain is not enclosed by a membrane. No sharp dividing line separates its inside from its outside. Yet it still maintains a chemical environment distinct from that of other areas. The scientists refer to it as a membraneless organelle.
Helping membrane proteins find their way to a cell’s surface is clearly one of TIGER’s functions, but there could be others. Dr. Mayr thinks that another function might be to provide a space where mRNAs can hibernate until they are needed by a cell. She gives the example of immune cells, which keep quiet in the body until they encounter a trigger, like a virus or bacteria. Then they spring to life and start making proteins that drive an immune response. Having mRNAs readily available might save the body valuable time in fighting infections.
She also thinks that these membraneless organelles might be present throughout the cytoplasm. If that’s the case, then scientists will have to revise another common conception about cells — that they are more or less squishy bags filled with water. The existence of regions like the TIGER domain implies that the cytoplasm itself has distinct compartments that aren’t defined by membranes.
“Right now we don’t see these compartments,” Dr. Mayr says. “Then again, nobody knew that TIGER was there until we looked for it.”