
Graduate training: I was trained towards my PhD with Dr. Gutian Xiao at University of Pittsburgh School of Medicine. There I made mechanistic discoveries related to a role of autophagy and NF-kB in inflammation and cancer. My work was the first to provide evidence that an HSP90 client can be degraded by a mechanism different from the ubiquitin-proteasome pathway and to establish a molecular link among HSP90, NF-kB and autophagy. I also investigated the regulation of HTLV-I oncoprotein Tax by PDZ-LIM domain-containing protein (PDLIM2); specifically I identified PDLIM2 as a negative regulator of HTLV-I oncoprotein Tax. I found that the counterbalance between HTLV-I Tax and PDLIM2 determined the outcome of the HTLV-I infection. Furthermore, I defined the molecular determinants of PDLIM2 in suppressing Tax mediated tumorigenesis. By dissecting the functional sequences within PDLIM2 my work provided insights into the novel oncogenic mechanisms in HTLV-I leukemogenicity and cancer health disparities, and suggested potential therapeutic approaches against Adult T-cell Leukemia.
Postdoctoral research: With my molecular biology and biochemistry credentials assured, I was recruited to be a member of Dr. Gabriela Chiosis’ laboratory at MSKCC after defending my PhD thesis. The interdisciplinary environment of the lab fostered my interest in chemical biology and translational research. My research moved to the understanding of how molecular chaperones aid in malignant transformation. I also developed a keen interest in the translation of mechanistic findings into novel therapies.
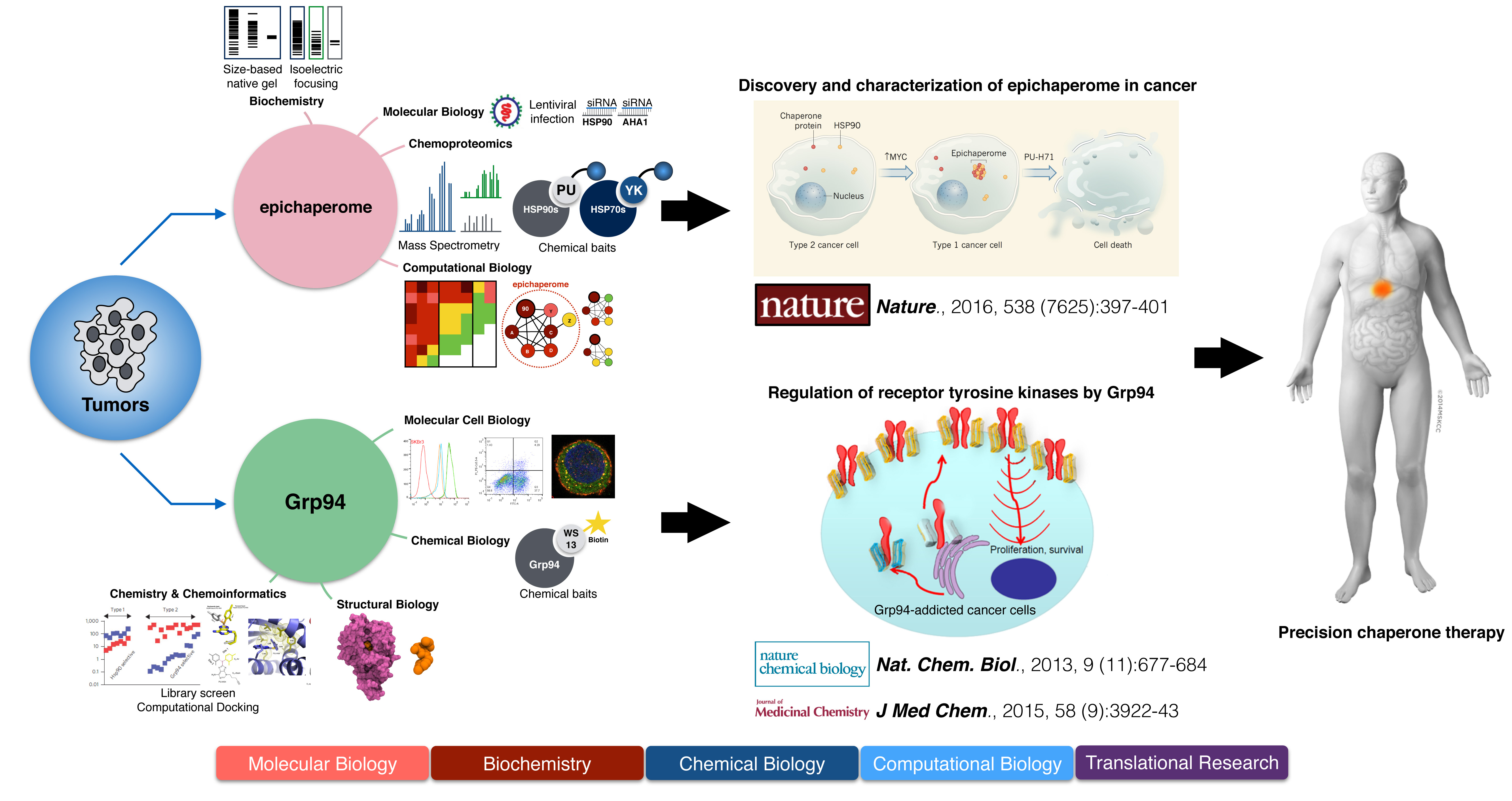
My first project had GRP94, the endoplasmic reticulum (ER) HSP90 paralog, the focus of my research. HSP90, the most abundant molecular chaperone in human cells has four paralogs. Whereas much work was dedicated to the study of the cytosolic HSP90 paralogs, HSP90α/β, little is known on molecular mechanisms associated with GRP94 in cancer. There is no GRP94 homolog in a genetically tractable organism such as yeast, and studies using mutant cell lines and gene-deficient mouse studies have been of limited use. To address this limitation, my lab created a variety of reagents and assays to enable the study of GRP94 in native tumors. I used these tools to find an unexpected role for GRP94 in cancer – cancer cells that activated signaling pathways through overexpression of receptor tyrosine kinases, translocated GRP94 to the plasma membrane and used it to regulate kinase activity at this location. My data indicated that only in cells with HER2 overexpression, the chaperoning function of GRP94 was vital for proper HER2 function. I also demonstrated that pharmacological inhibition of GRP94 in these cells was sufficient to destabilize membrane HER2, inhibit its signaling properties and target HER2 towards a degradative pathway. These findings were the first to implicate GRP94 in regulating oncogenic signal transduction at the plasma membrane. These effects were previously unknown and unanticipated although HER2 is a widely-studied HSP90 client protein. Also of importance, this work provided proof for the integration of the HSP90 chaperone system - when faced with an ‘over-demand’ from the proteome, in this case HER2 overproduction and enhanced HER2 signaling, the cancer cell brought in distinct HSP90 paralogs to cooperatively regulate HER2.
Over the past two years I made significant contributions to another major project in the lab. We found that cancer cells regulate proteome stress not only by chaperone overexpression, but rather by a re-wiring of the chaperone units. For example, in large proteome imbalances produced by MYC activation, we identified that cancer cells re-wire the majority of the chaperone units into large chaperone networks (Nature 2016). These networks, created by a change in the cellular milieu rather than defects in chaperone structure or expression, we term the epichaperome. These networks protect certain cancers against cell death during their transformation into malignancies. While chaperone units are abundant and expressed virtually in every cell in the human body, epichaperome networks are unique to cancer cells. When we looked for proteome stress that could create epichaperome, we recognized that cancer cells sensitive to chaperone inhibition had in common a MYC transcriptional signature. These cancer cells had indeed higher MYC transcriptional activity; and by turning MYC on, we could assemble the chaperome into epichaperome networks and make cancer cells sensitive to HSP90 inhibition. Conversely, by turning MYC off, we could dis-assemble the chaperome and make cancer cells insensitive.
In addition to providing a novel mechanism of tumor regulation, this work has two other major implications. First, it provides a means for precision chaperone therapy based not on genetics but rather on protein networks. We find that over half of cancers, irrespective of genetic defect, type, site or origin use epichaperome networks to regulate alterations in the proteome and maintain cellular survival. The finding provided a breaking-through point-of-view that revolutionized traditional paradigm that narrowly defines the drug selectivity by protein over-expression or mutations. We demonstrated that for a ubiquitously expressed chaperone (i.e. HSP90), drug sensitivity is not solely associated with its abundance, but rather determined by the binding affinity and interacting chaperome. Such discovery will prompt scientists to adopt a more dynamic view towards investigating cancer, rather than relying on static biomarkers. Second, it provides a means for building sequential combination therapies that incorporate HSP90 agents. A clinical study built on these findings is to start at MSKCC later this year.