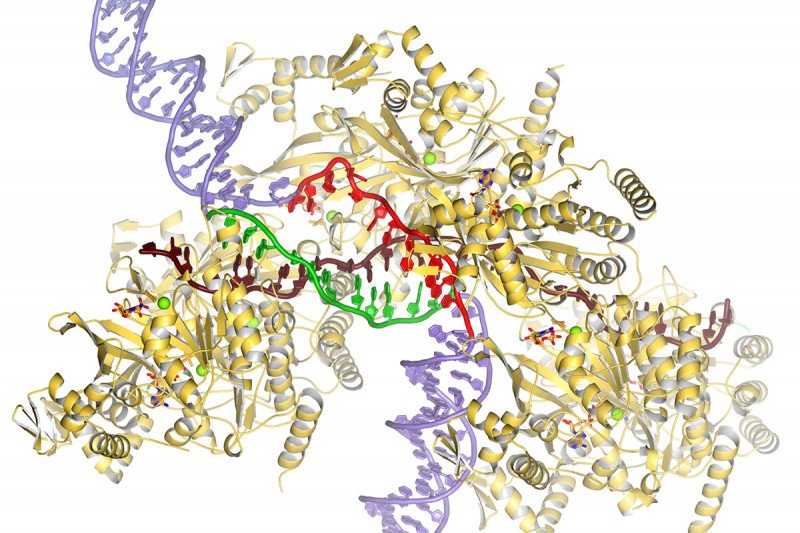
This illustration shows proteins and DNA in the process of DNA repair. RecA proteins (gold) cradle a single-stranded DNA (brown) that searches for homology with a DNA template (purple). The template DNA is opened up, and one of its strands forms new double-stranded DNA (green and brown) while the other is sequestered away (red).
A landmark in cancer research was the discovery, in the early 1990s, of two cancer predisposition genes — BRCA1 and BRCA2. When mutated, these genes increase a person’s risk of developing several forms of cancer, including breast, ovarian, and prostate cancers. Since then, researchers around the world have been studying these genes and the proteins made from them to learn exactly how they increase the risk of cancer.
Many crucial breakthroughs in scientists’ understanding of this topic have come from research conducted at Memorial Sloan Kettering. In the late 1990s, Sloan Kettering Institute molecular biologist Maria Jasin showed that the BRCA2 protein was necessary for repairing a type of DNA damage called a double-strand break. When BRCA2 is mutated, it can’t repair this damage well, and cancer is often the result.
In 2002, SKI structural biologist Nikola Pavletich determined the structure of the BRCA protein and showed that it binds to DNA. In a follow-up paper published in 2005, Dr. Pavletich showed that BRCA2 is required for DNA repair because it activates another protein, called Rad51, which is the actual machine that repairs double-strand breaks. In 2008, he showed how the bacterial version of Rad51, called RecA, starts the repair process by binding to one strand of the broken DNA.
Now, more than 12 years later, Dr. Pavletich has added yet another piece to the puzzle. In a report published October 14 in the journal Nature, he and his colleagues, including senior research scientist Haijuan Yang, describe the mechanism by which RecA and the broken DNA strand it carries search for the correct segment of a nearby DNA molecule to use as a repair template.
The findings cap a nearly 20-year quest, the pace of which has accelerated in recent years thanks to a new technology called cryo-electron microscopy (cryo-EM).
Searching for a Match
Because DNA is so fundamental to life, cells have evolved a variety of means to preserve its integrity. When DNA is damaged — say by UV light or x-rays — there are several ways a cell can attempt to repair it. The most careful, error-free way is called homologous recombination. In this type of repair, a cell finds a segment of DNA on an intact chromosome that matches the broken region of the other and uses that as a template to fill in the gap. (Chromosomes come in pairs, one from mom, one from dad; these chromosomes are said to be ‘homologous.’)
From decades of work, it was clear that a single strand of broken DNA finds homologous DNA with the help of a protein called RecA. RecA and single-stranded DNA form a structure called a pre-synaptic filament. The filament binds to double-strand DNA, opens it up to expose its complementary strands, and then searches for homology along one of those strands.
“The question we didn’t understand was how it’s searching for homology between the single-stranded DNA that it’s carrying and the double-stranded DNA that it binds to,” Dr. Pavletich says.
To investigate that question, they turned to cryo-EM. This relatively new form of microscopy, whose developers won the 2017 Nobel Prize in Chemistry, allows scientists to visualize the fine-grained structure of biological molecules at unparalleled resolution. MSK acquired a cryo-EM machine in 2017.
Before the advent of cryo-EM, the main way that biologists determined structures of proteins and other molecules was x-ray crystallography, a painstaking and time-consuming effort.
“We’ve spent a long time with crystallography trying to address these issues and it just did not work out,” Dr. Pavletich says. “Cryo-EM makes it much easier.”
Part of the problem is that to really understand the mechanism, they needed to capture what the RecA protein looks like at various points during the DNA repair process; with x-ray crystallography, they were really only able to look at one point in time.
With cryo-EM, they are able to look at thousands and thousands of RecA proteins bound to DNA at various times points and from these many images piece together the entire sequence.
“What we were able to determine is how RecA opens the double-stranded DNA and how one of the DNA strands is sequestered at a second secondary site in the RecA protein,” Dr. Pavletich says.
“This binding leaves the other DNA strand flipping around and hitting the single-stranded DNA that the filament was carrying,” he adds. “If there is homology, it stays there. If there are no homology it continues to float around.”
Even if the team had had several x-ray crystal structures to work with, they wouldn’t have been able to glean what they were able to using a million RecA-DNA particles. “That’s the power of cryo-EM,” he says. “It took us 12 years to solve this, but if we had had cryo-EM in 2008 then it would have been much faster.”
For the Love of Discovery
Homologous recombination is not only important in fixing mutations that can lead to cancer. It’s also how genetic diversity is generated during the formation of sperm and egg: maternal and paternal chromosomes break along their lengths and swap segments before being repaired through homologous recombination. This genetic diversity is why you and your siblings look similar but not identical; you each got a different combination of maternal and paternal chromosomes.
Homologous recombination is also how the genome-editing tool CRISPR works. This technology, whose developers won the 2020 Nobel Prize in Chemistry, relies on the introduction of DNA double-strand breaks at specific locations in the genome and repair through homologous recombination.
Dr. Pavletich says he hopes the new insights into homologous recombination will ultimately help improve cancer care, as past discoveries have done. But he says that’s not the primary goal of basic science research.
“As scientists, we do what we do because we love the gratification of solving a problem,” he says. “And homologous recombination is one of those really longstanding and important biological problems to understand. So it feels really good to be able to make this contribution to science.”